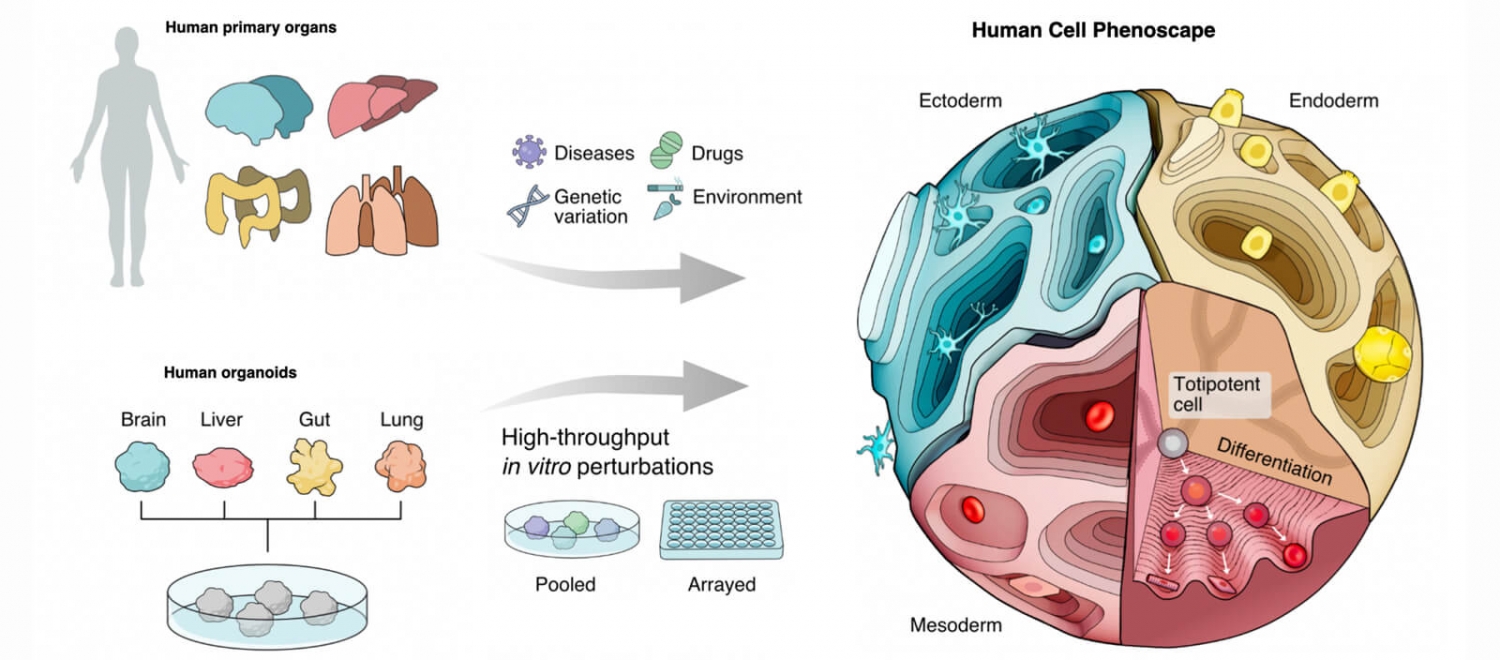
By leveraging emerging experimental and computational techniques, scientists suggest that we can gain valuable insight into the causal relationships between perturbation and cell state, potentially providing mechanistic insight of development or identifying drug targets.
Scientists envision a future where we can predict a cell’s future state. What if we knew how changes in a cell’s environment affected cellular response? By leveraging emerging experimental and computational techniques, scientists suggest that we can gain such valuable insight into the causal relationships between perturbation and cell state, potentially providing mechanistic insight of development or identifying drug targets. In a Perspective paper published in the journal Science, scientists from IHB and ETH Zürich’s Dept of Biosystems explore this potential.
Current efforts worldwide are rapidly progressing to comprehensively understand the complex and diverse array of cells constituting the human body. These endeavors aim to categorize cell types based on a multitude of molecular features encompassing aspects of the transcriptome, epigenome, proteome, and developmental lineage. In other words, they’re creating detailed maps, called cell atlases, of different categories of cell types, or phenotypes, based on molecular signatures.
However, cells are not static and the complexity of human cell types extends beyond molecular signatures. Cells exhibit diverse states that are influenced not only by their developmental history and current molecular profile but also by their responses to environmental changes or perturbations. Perturbations, ranging from genetic alterations to chemical stimuli, can induce a spectrum of phenotypic changes and lead to reversible or irreversible shifts in cellular identity.
While single-cell genomic tissue atlases have made significant impacts in biomedical research, they are limited as physiological snapshots and cannot reveal causal factors of development or predict unseen perturbed cell states. This is where complex three-dimensional human cell culture models, such as organoids, can fill the gap as they can mimic key aspects of human physiology and are compatible with high-throughput perturbation experiments and multimodal phenotyping.
“It’s an exciting time for computational biology. With new measurement modalities and organoid technologies we can acquire rich data at unprecedented scales. For the first time, this might allow us to predictively model complex biology.”
Jonas Fleck
Postdoctoral Researcher, Developmental Systems and Computational Biology group, IHB

Postdoctoral researcher and first author Jonas Fleck (IHB) and co-authors Gray Camp (IHB), Barbara Treutlein (D-BSSE, ETH Zürich) envision that perturbing human model systems at scale will help us better understand cell types by enabling us to chart the human cell phenoscape, or the totality of the human-cell phenotypic landscape. However, with infinitely many possible perturbations, mapping out the entire phenoscape seems daunting. Predictive modeling based on machine- and deep- learning offers an efficient solution by not only extrapolating to unseen contexts but also quantifying areas that are sufficiently charted and those that still need exploration.
The ultimate goal remains to develop predictive models capable of anticipating the effects of perturbations on cell phenotypes, thereby charting the expansive phenoscape with increasing accuracy and efficiency. Such models, driven by large and diverse datasets, could guide future research endeavors to understand health and disease states and aid in the identification of novel therapeutic targets.
The totality of the human-cell phenotypic landscape (“phenoscape”) can be simplified and visualized as a topological and spherical map, where the totipotent cells at the core differentiate toward the surface topology. Blue represents ectoderm, orange represents endoderm and red represents mesoderm. Illustration by Melanie Lee, Nanoclustering. A version of this image was published in Fleck et al. 2023. Science
Journal paper is available open access: Fleck JS,Camp JG, Treutlein B. 2023. What is a cell type? Science
Read the full paper: https://www.science.org/doi/10.1126/science.adf6162