The success of IHB owes much to the wonderful and inspiring people who work in its labs. We are featuring Bruno Gjeta, who is the first student to receive his PhD at IHB and works in the Developmental Systems and Computational Biology team.

“Since childhood, I have always been curious about the complexity of the realities around me.”
Bruno Gjeta
Research Fellow
Show profile
Since childhood, I have always been curious about the complexity of the realities around me. I wondered how things work and dreamt of being a scientist. These feelings got reinforced during adolescence after my Retinitis Pigmentosa diagnosis. From then on I aspired to fuse my curiosity with the purpose of providing therapeutic solutions to patients who, just like me, did not have any. This initiated me to biotechnology in my early 20s and eventually led me to where I am today.
IHB is an interdisciplinary entity which strives at improving our understanding of human biology as well as disease phenotypes by leveraging innovative technologies. I wanted to join IHB for the opportunity to collaborate and learn from so many great scientists and together advance fundamental research as well as translational solutions. As a personal achievement, I am proud to say that I was the first PhD graduate of both IHB and Gray Camp’s lab.
My research focused on investigating human disease progression and therapeutic potential using single-cell technologies and organoid models. The goal is to identify key principles that drive the progression of a particular disease or undesired effect of a treatment. In this endeavor, we aim at gaining a set of more specific molecular features that could help detect the disease state early on as well as understanding which levers we can pull when trying to prevent progression into harmful phenotypes. The hope is that this knowledge could translate into therapeutic targets in areas with as yet unmet clinical needs.
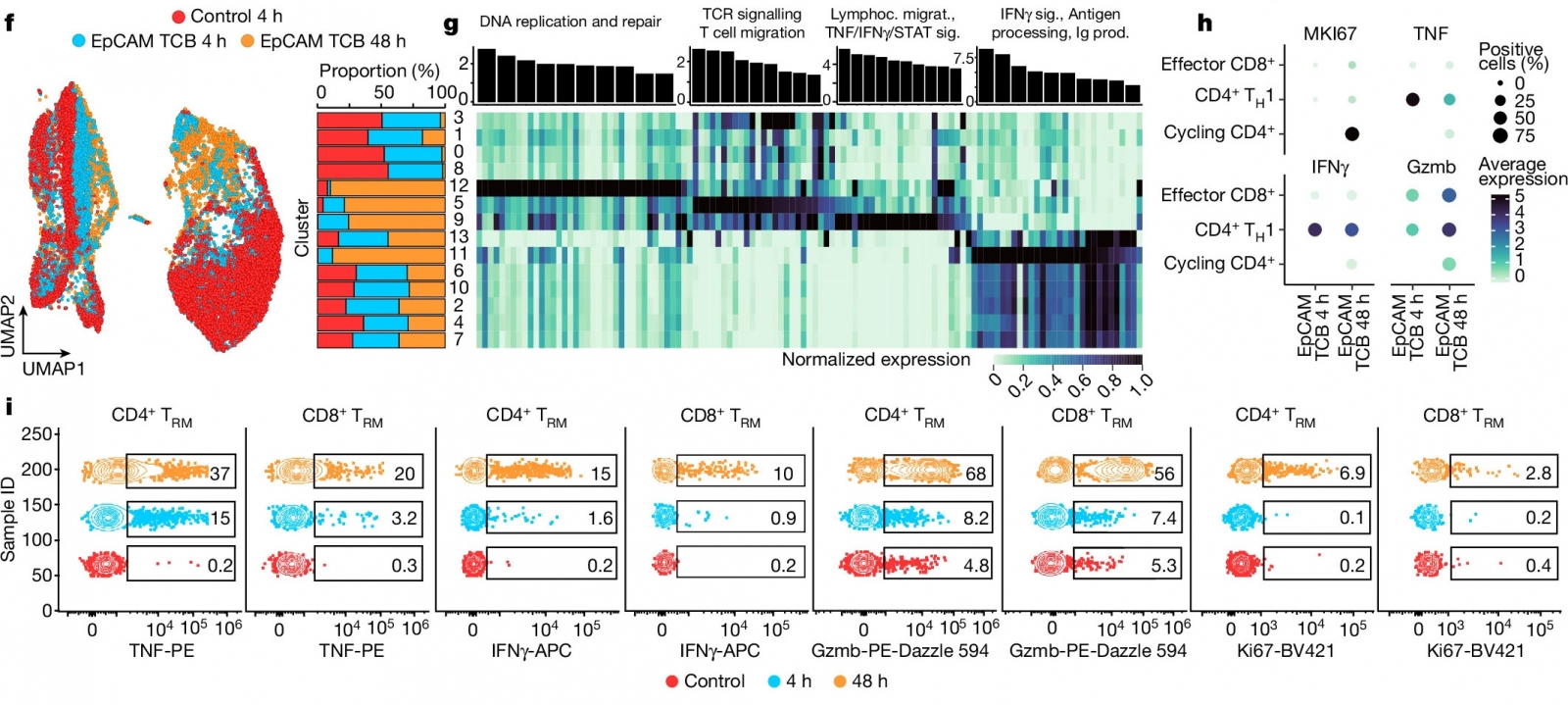
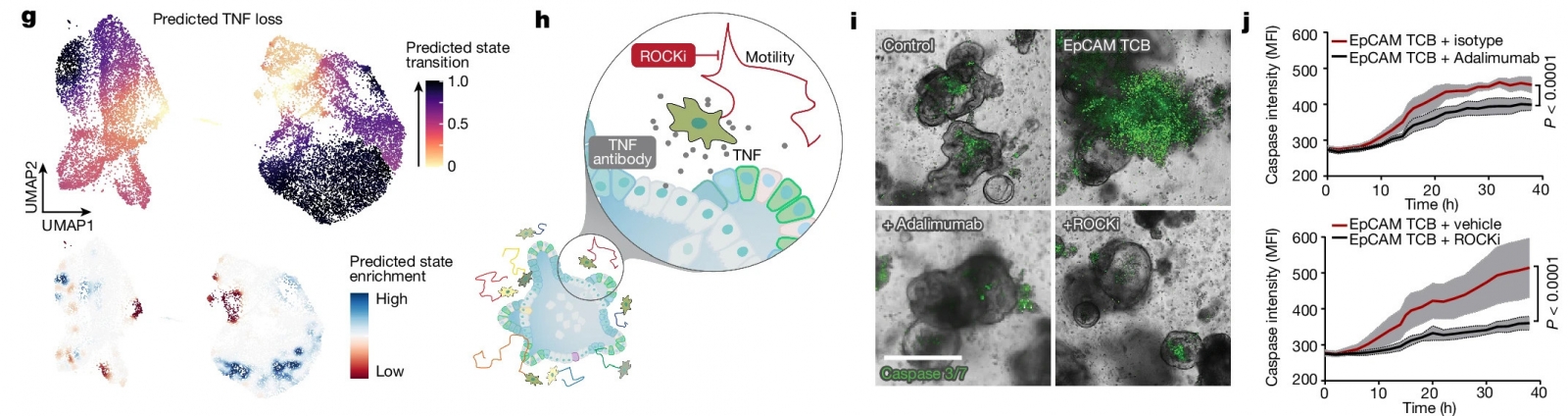
My expertise in human biology and computational analysis contribute to identifying, understanding and communicating scientific insights that help push IHB’s collaborative projects. In one recent work, a collaboration between three different teams at IHB, we described how interactions among tissue-resident gut immune cell populations contribute to cytotoxicity in response to cancer biologics. We identified and described specific contributors to the cytotoxic activation and validated strategies to mitigate it which provide an inroad to manage clinical side effects when developing new therapeutics.
My time at IHB has been valuable in helping me understand what I enjoy and where I can grow further. This will guide me in pursuing opportunities that are more aligned with my long-term goals.
The IHB community is quite diverse with people coming from lots of different countries and backgrounds. This opens the door for very interesting and fun interactions over a beer by the Rhein.
Recaldin T*, Steinacher L*, Gjeta B*, Harter MF*, …, Cabon L^, Camp JG^, Gjvorevski N^ (2024) Human organoids with an autologous tissue-resident immune compartment. Nature.
*lead authors, ^senior corresponding authors
DOI: 10.1038/s41586-024-07791-5